Lumpy Skin Disease (LSD) is a devastating viral disease of cattle that threatens food security and rural livelihoods across Africa. While the disease originated on the continent, very little genomic data has been available from West and Central Africa, leaving gaps in our understanding of how the virus spreads and evolves.
A new study by our researchers has helped close this gap by sequencing and analysing LSDV strains from Nigeria, Cameroon, and the Benin Republic. The findings are significant for both science and farmers:
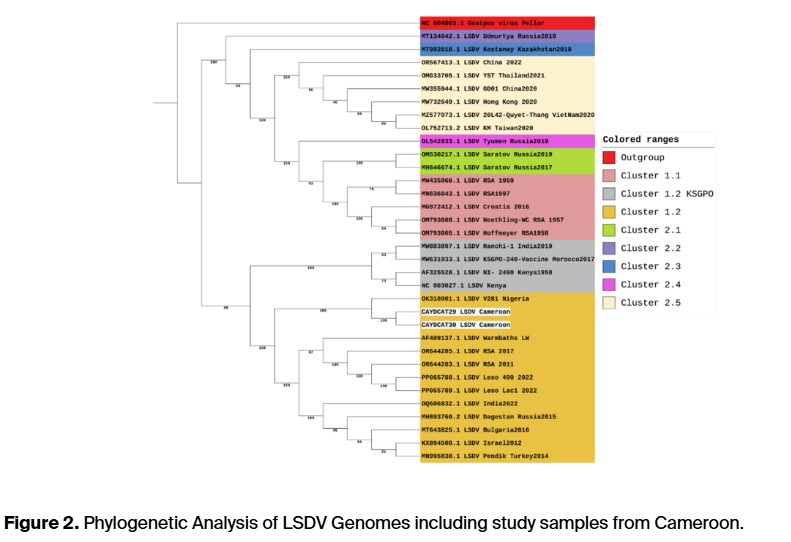
A new regional sub-lineage was identified: The strains clustered within the known 1.2 lineage but formed a distinct West/Central African branch, showing the importance of local genomic studies.
Mutations with vaccine implications: Genetic changes were found in viral proteins linked to immune evasion and virulence, raising concerns that current vaccines may not provide full protection in this region.
First evidence of flies as carriers: LSDV was detected in flesh flies (Sarcophaga spp.), marking the first report of this insect as a potential vector for the virus.
Genomic stability with regional uniqueness: The Nigerian and Cameroonian strains showed strong similarity to each other but noticeable differences compared to strains from Europe, Asia, and Southern Africa.

Why this matters:
For countries like Nigeria, where cattle are central to food security, trade, and rural livelihoods, these insights are practical. They highlight the urgent need for:
Improved vaccines tailored to African strains.
Continuous genomic surveillance to detect emerging variants early.
Vector control strategies are given the role of insects in spreading the disease.
This study is a clear reminder that African-led science is essential for solving African challenges. By understanding the genetics of LSDV where it matters most, the region is better positioned to protect farmers, strengthen animal health, and secure food systems.
Read full Publication here: https://www.mdpi.com/2076-0817/14/9/922

